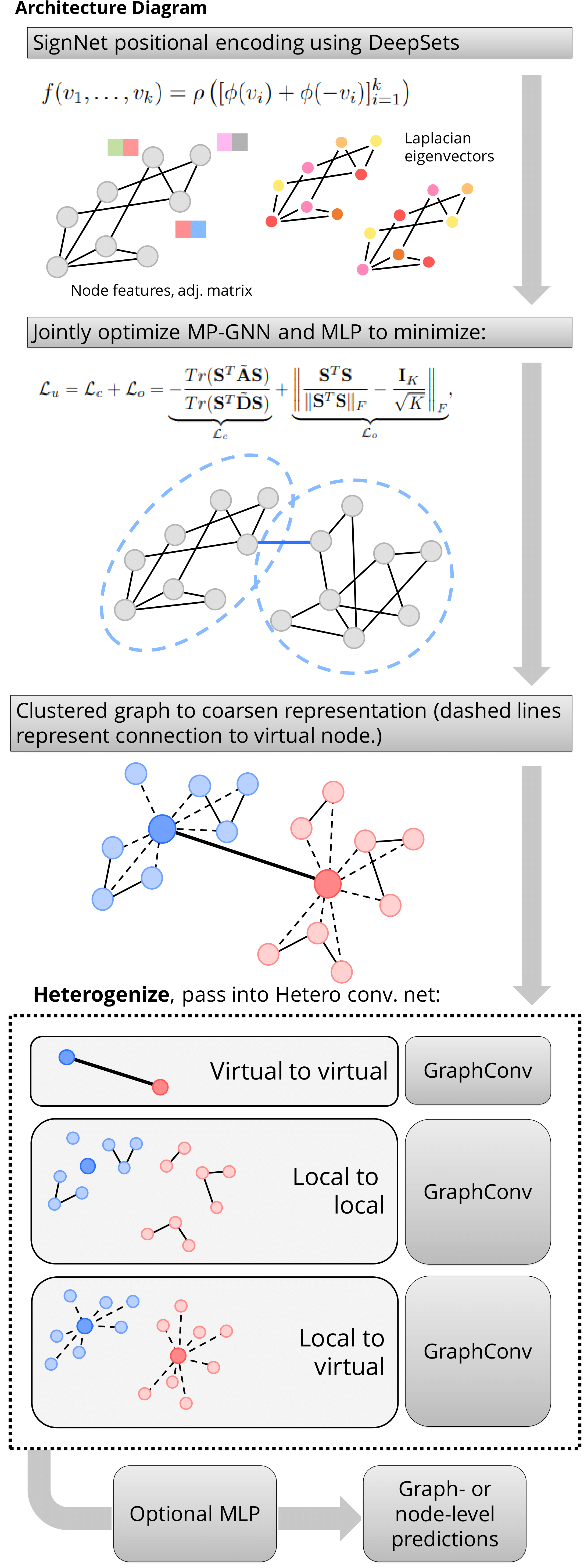
Architecture
Our model is composed of the following components:
-
SignNet Positional Encoder
- In the architecture we determined to use SignNet to compute the positional encoding for each graph. We pre-computed the top 50 eigenvalues and corresponding eigenvectors of the laplacian matrix L of each graph. Then we used the com- puted top 50 eigenvectors (Nx50) as the input for the SignNet.
-
Spectral clustering
- We trained a GNN model using pytorch-geometric and used the loss functions as the sum of minCUT Loss and orthogonal Loss to compute the cluster assignment for each graphs. We trained for each graph for about 50 iterations with a early stop threshold of loss decrease as 0.001. With the trained model we get the cluster assignments which we used in the next step to create virtual nodes and construct the heterogenous graph dataset.
-
Heterogeneous graph
- From the original graphs after we trained and computed the clusters we will add one new virtual node for each of the clus- ter. For K clusters, where K is a arbitrary number, we will add K new virtual nodes. For each virtual node, we will add M new edges between all the nodes in one cluster and the new virtual node, where M is the number of the nodes in a clus- ter. After virtual nodes connect the local nodes, we will fully connect all the virtual nodes. To address the different proper- ties of the edges from local nodes to local nodes, the edges from local nodes to virtual nodes and the edges from virtual nodes to virtual nodes, we applied the idea of Heterogenous Graph Neural Network by using different message passing convolution layers for different types of edges.